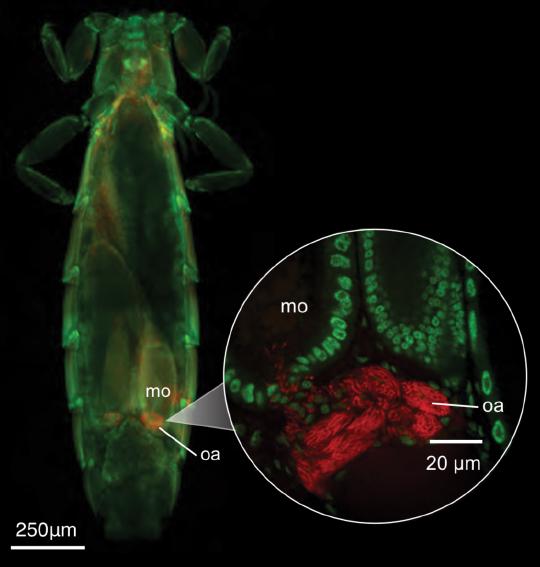
The effective population size (Ne) of an organism is expected to be generally proportional to the total number of individuals in a population. In parasites, we might expect the effective population size to be proportional to host population size and host body size, because both are expected to increase the number of parasite individuals. However, among other factors, parasite populations are sometimes so extremely subdivided that high levels of inbreeding may distort these predicted relationships. Here, we used whole-genome sequence data from dove parasites (71 feather louse species of the genus Columbicola) and phylogenetic comparative methods to study the relationship between parasite effective population size and host population size and body size. We found that parasite effective population size is largely explained by host body size but not host population size. These results suggest the potential local population size (infrapopulation or deme size) is more predictive of the long-term effective population size of parasites than is the total number of potential parasite infrapopulations (i.e., host individuals).
Doña, J., Johnson, KP.,
Evol Lett,
2023
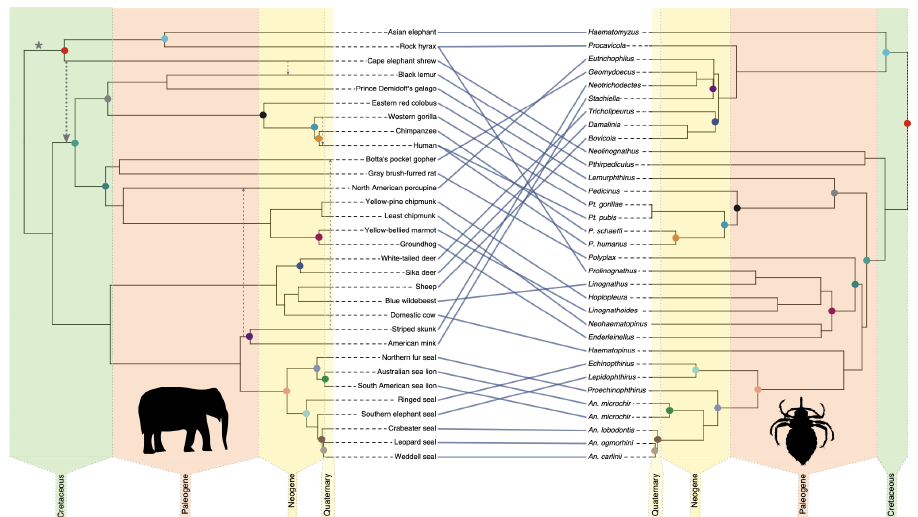
Mammals host a wide diversity of parasites. Lice, comprising more than 5,000 species, are one group of ectoparasites whose major lineages have a somewhat patchwork distribution across the major groups of mammals. Here we explored patterns in the diversification of mammalian lice by reconstructing a higher-level phylogeny of these lice, leveraging whole genome sequence reads to assemble single-copy orthologue genes across the genome. The evolutionary tree of lice indicated that three of the major lineages of placental mammal lice had a single common ancestor. Comparisons of this parasite phylogeny with that for their mammalian hosts indicated that the common ancestor of elephants, elephant shrews and hyraxes (that is, Afrotheria) was the ancestral host of this group of lice. Other groups of placental mammals obtained their lice via host-switching out of these Afrotherian ancestors. In addition, reconstructions of the ancestral host group (bird versus mammal) for all parasitic lice sup- ported an avian ancestral host, indicating that the ancestor of Afrotheria acquired these parasites via host-switching from an ancient avian host. These results shed new light on the long-standing question of why the major groups of parasitic lice are not uniformly distributed across mammals and reveal the origins of mammalian lice.
Johnson, KP., Matthee, C., Doña, J
Nat Ecol Evol,
2022
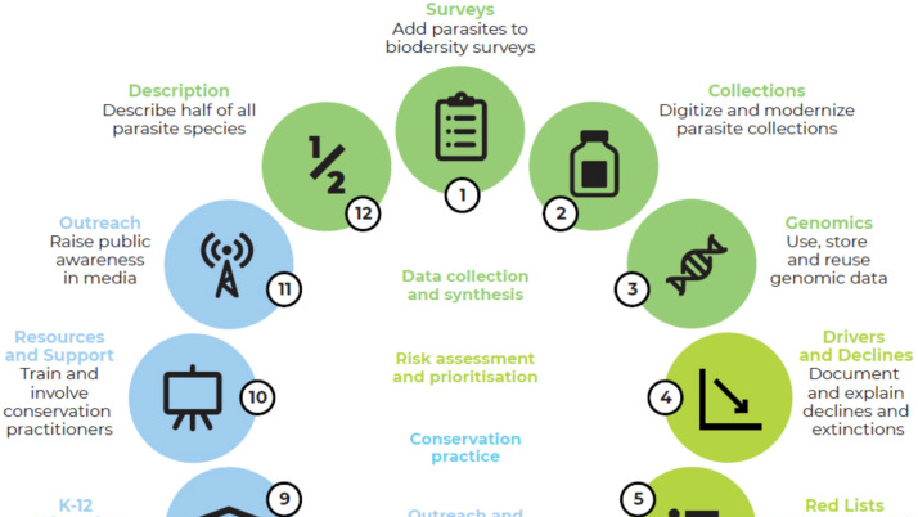
Found throughout the tree of life and in every ecosystem, parasites are some of the most diverse, ecologically important animals on Earth—but in almost all cases, the least protected by wildlife or ecosystem conservation efforts. For decades, ecologists have been calling for research to understand parasites’ important ecological role, and increasingly, to protect as many species from extinction as possible. However, most conservationists still work within priority systems for funding and effort that exclude or ignore parasites, or treat parasites as an obstacle to be overcome. Our working group identified 12 goals for the next decade that could advance parasite biodiversity conservation through an ambitious mix of research, advocacy, and management.
Carlson, CJ., Hopkins, S., Bell, KC., Doña, J., Godfrey, SS., Kwak, M., Lafferty, KD., Moir, ML., Speer, KA., Strona, G., Torchin, M., Wood, C
Biol. Conserv,
2020